Machine Learning-Based Invariant Feature Engineering (LIFE) | |||||
|
|||||
Summary ^Symmetry refers to properties that remain invariant upon mathematical transformations, yet it remains unexplored in biology and medicine. We set to explore symmetry relationships in gene expression to distinguish between healthy and disease states. We hypothesize that there are relationships between gene expressions that remain invariant across individuals displaying the same biological phenotype. Our Gene Expression Symmetry Hypothesis (GESH) posits that the invariant nature of phenotypic traits in cells is defined by a set of genes exhibiting specific symmetric expression relationships. We deployed a hybrid machine learning approach implemented with two symmetric invariant feature functions (IFFs) to identify Invariant Feature Genes (IFGs), which are gene pairs whose IFF single-value outputs remain invariant across individual samples in each phenotype. Our multiclass classification results across the transcriptomes of 25 normal organs, 25 cancer types, and blood samples from 4 different types of neurodegenerative diseases identified unique fingerprints. We constructed networks from IFGs and found that cancer IF-Nets hubs were enriched with approved and clinical trial drugs, highlighting “symmetry breaking” as a novel treatment approach. 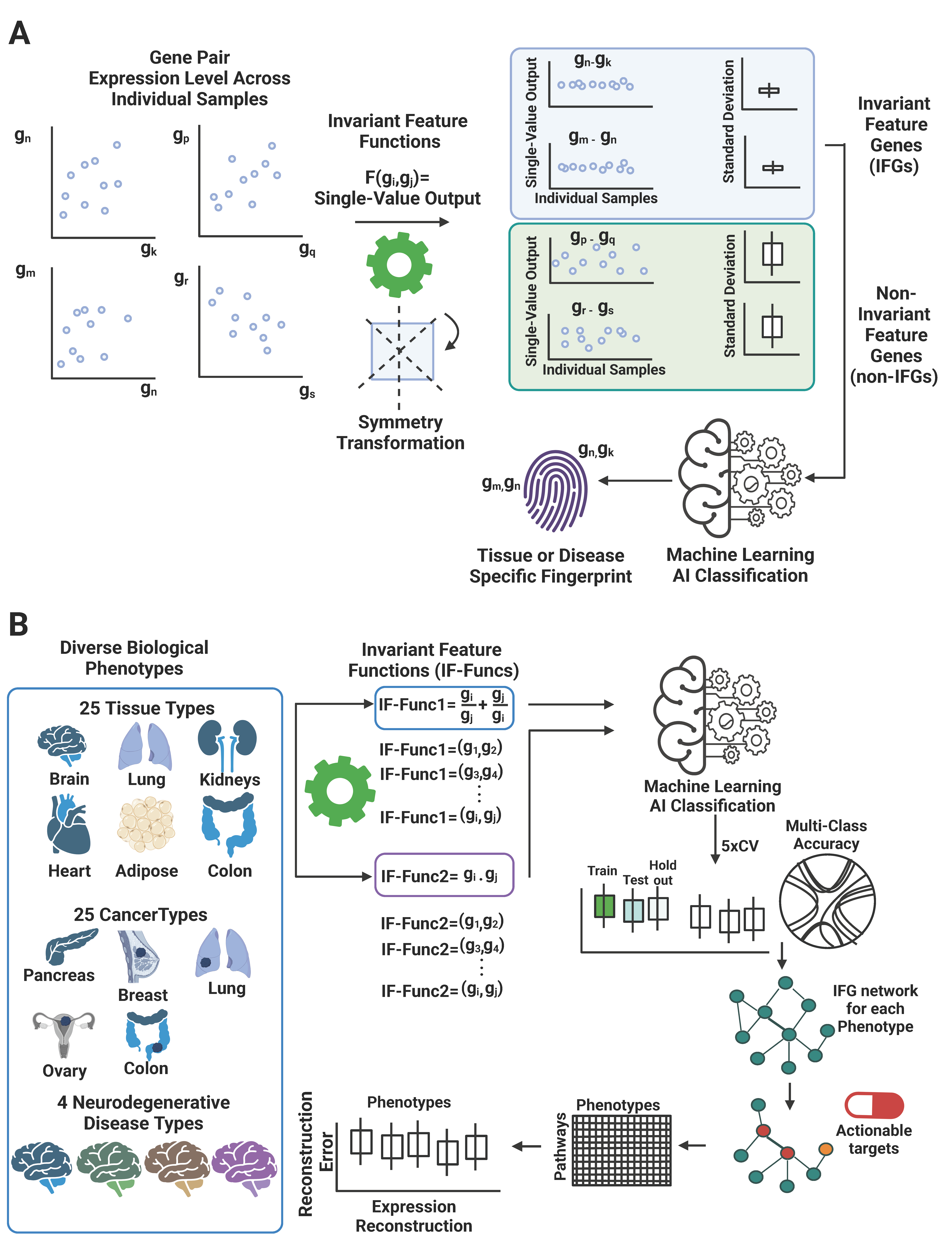
Symmetric gene expression relationships define a phenotypic trait. Results ^
Explore drug target networks of IFF1 and IFF2 here. Download ^comming soon. Support ^comming soon Citation ^
Manuscript in preparation. |